Division of Integrative Biomolecular Function
Numerous organisms have established universal and unique biologically regulatory systems, and biologically and ecologically interact among them. Such biodiversity might have been evolved via a wide range of selective pressures and interphyletic biological interaction. However, the underlying molecular mechanisms largely remain unknown. Our group has been investigating the essential principles and biological significances for the generation of the biodiversity and the ecological systems for animals, plants, and microbes as the molecular and genetic networks using multiple approaches, including molecular biology, cell biology, physiology, bioinformatics, and analytical chemistry.
Group Members
Division of Integrative Biomolecular Function
General Manager/ Executive Researcher
Honoo SATAKE
Senior Researcher
Tsuyoshi KAWADA
Executive Researcher
Toshio TAKAHASHI
Senior Researcher
Jun MURATA
Senior Researcher
Tomotsugu KOYAMA
Researcher
Tsubasa SAKAI
Researcher
Tomohiro OSUGI
Researcher
Akira SHIRAISHI
Researcher
Tatsuya YAMAMOTO
Senior Researcher
Shin MATSUBARA
Researcher
Yuta TAKASE
Appointed Researcher
Yoshiko MURATA
Technical Assistant
Shinji KIRIMOTO
Technical Assistant
Azumi WADA
Technical Assistant
Mika NOBUHARA
Technical Assistant
Hiromi TOYONAGA
Research Projects
1. Molecular mechanisms for signaling molecule and metabolizing enzyme-directed evolution and diversification
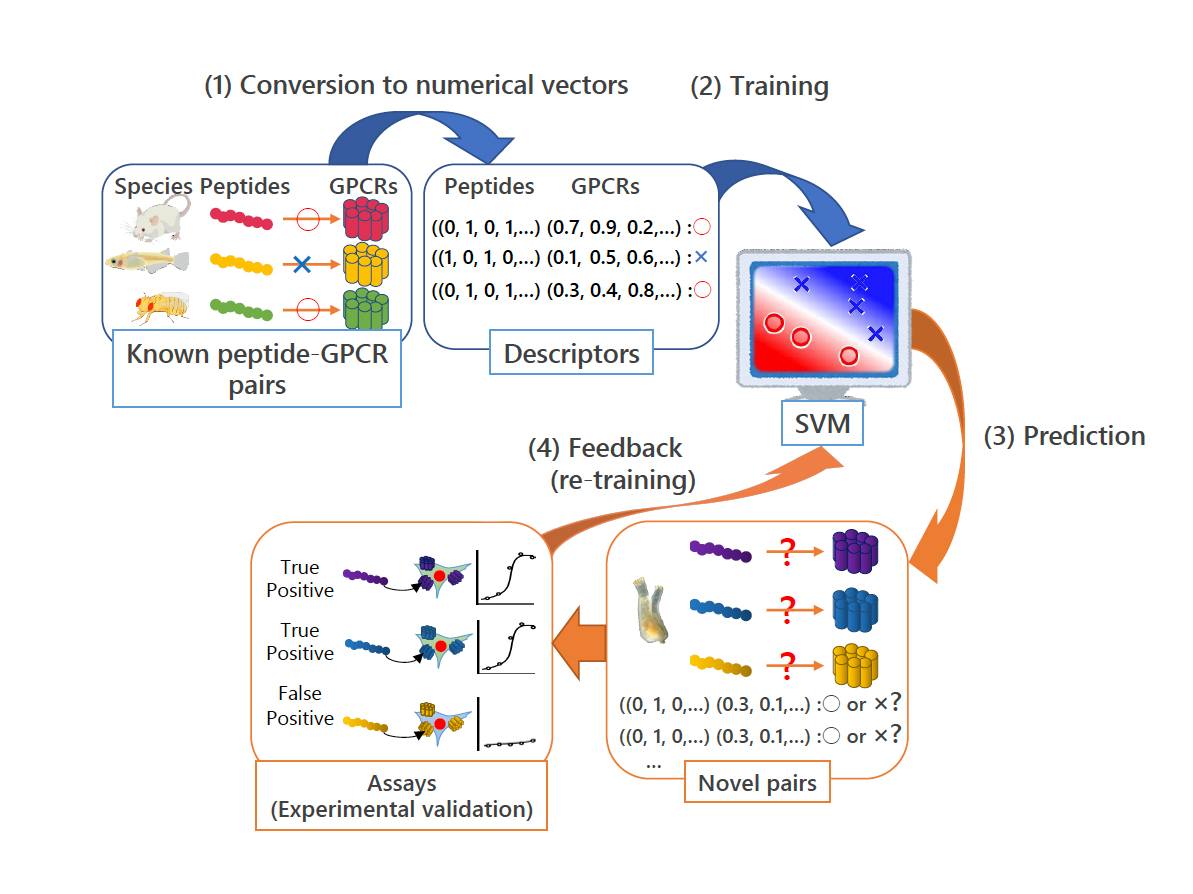
Neuropeptides and peptide hormones play pivotal roles in a variety of biological events, including reproduction, nuterient uptake, homeostasis, and learning. We have so far identified both homologous and novel neuropeptides and peptide hormones (more than 90% of C. intestinalis peptides) in the ascidian, Ciona intestinalis Type A (Ciona robusta). We have also elucidated the cognate receptors by a combination of machine learning-based prediction of peptide-receptor interactions and reverse-pharmacological methods (Fig. 1). We have also substantiated the regulation of signaling by Ciona-specific GnRH receptor paralogs via heterodimerization. This is the first report on GPCR heterodimerization among species-specific receptor paralogs.
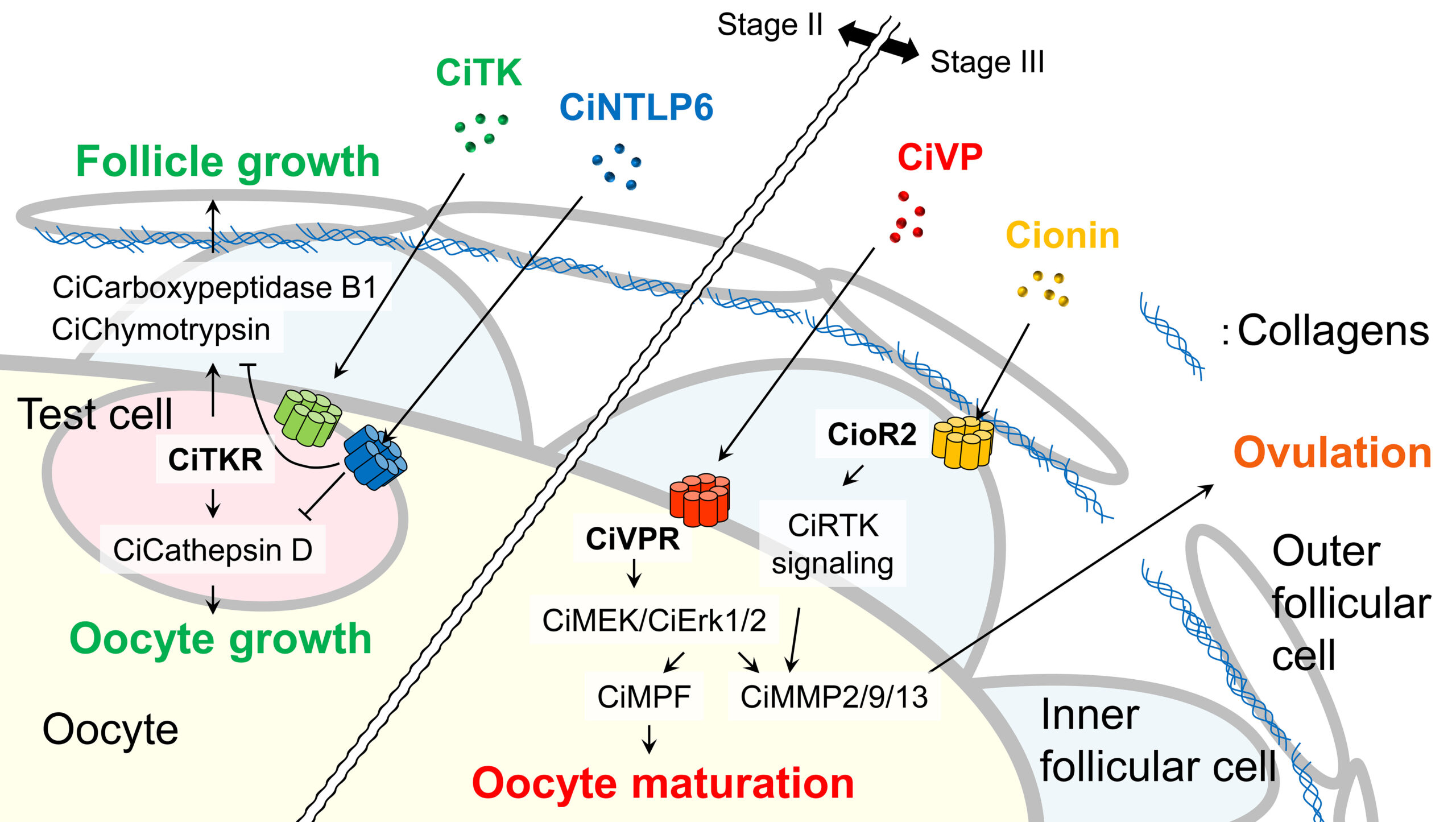
Moreover, we have demonstrated that a number of Ciona peptidergic neurons innervate the ovary, and investigated a novel ovarian follicle growth, maturation, and ovulation pathway in Ciona. CiTK, the Ciona tachykinin ortholog, has been shown to specifically up-regulate the late vitellogenic follicles via activation of several proteases such as cathepsin D, carboxypeptidase B1, chymotrypsin (Fig. 2). The CiTKergic follicle growth has been found to be down-regulated by a Ciona-specific neuropeptide, Ci-NTLP6 (Fig. 2). The Ciona vasopressin ortholog, CiVP, has been shown to enhance follicle maturation (germinal vesicle breakdown, GVBD) via activation of MPF and ovulation via upregulation of MMP. Cionin, the Ciona cholecystokinin ortholog has also been found to upregulate MMP (Fig. 2). These are the first report on the molecular mechanisms underlying the non-vertebrate chordate follicle development and on biological roles for these peptide families in the reproductive processes.
In vertebrate endocrine systems, the hypothalamic-pituitary-gonad (HPG) axis regulates the reproductive functions via induction of gonadotropins from the pituitary by the hypothalamic peptide hormone, GnRH. In contrast, Ciona has not been endowed with a hypothalamus or pitutary, and GnRHs are not involved in the regulation of the follicle development. Consequently, our studies have verified the common and species-specific reproductive processes among chordates. We have now been investigating the Ciona reproductive functions and evolutionary processes of the follicle development throughout chordates in greater details.
2. Signaling pathways for intestinal stem cell maintenance via non-neuronal acetylcholine receptors
The small intestine is composed of crypt and villus structures (Fig. 1). Villi are specialized for digestion and absorption and crypts project on the outside of villi. The small intestinal epithelium is largely renewed every 3 to 5 days. Villi continuously shed differentiated cells from their tips, and these losses are replenished by intestinal stem cells (ISCs) located in the crypt bottom. The mouse small intestinal crypts form organoids that mimic the intestinal epithelium when cultured in three-dimensional Matrigel. The organoids are self-organizing structures, referred to as the “mini-gut” owing to the presence of the various differentiated lineages, undifferentiated, and stem cells present in a normal small intestinal epithelium and lack nerve and immune cells.
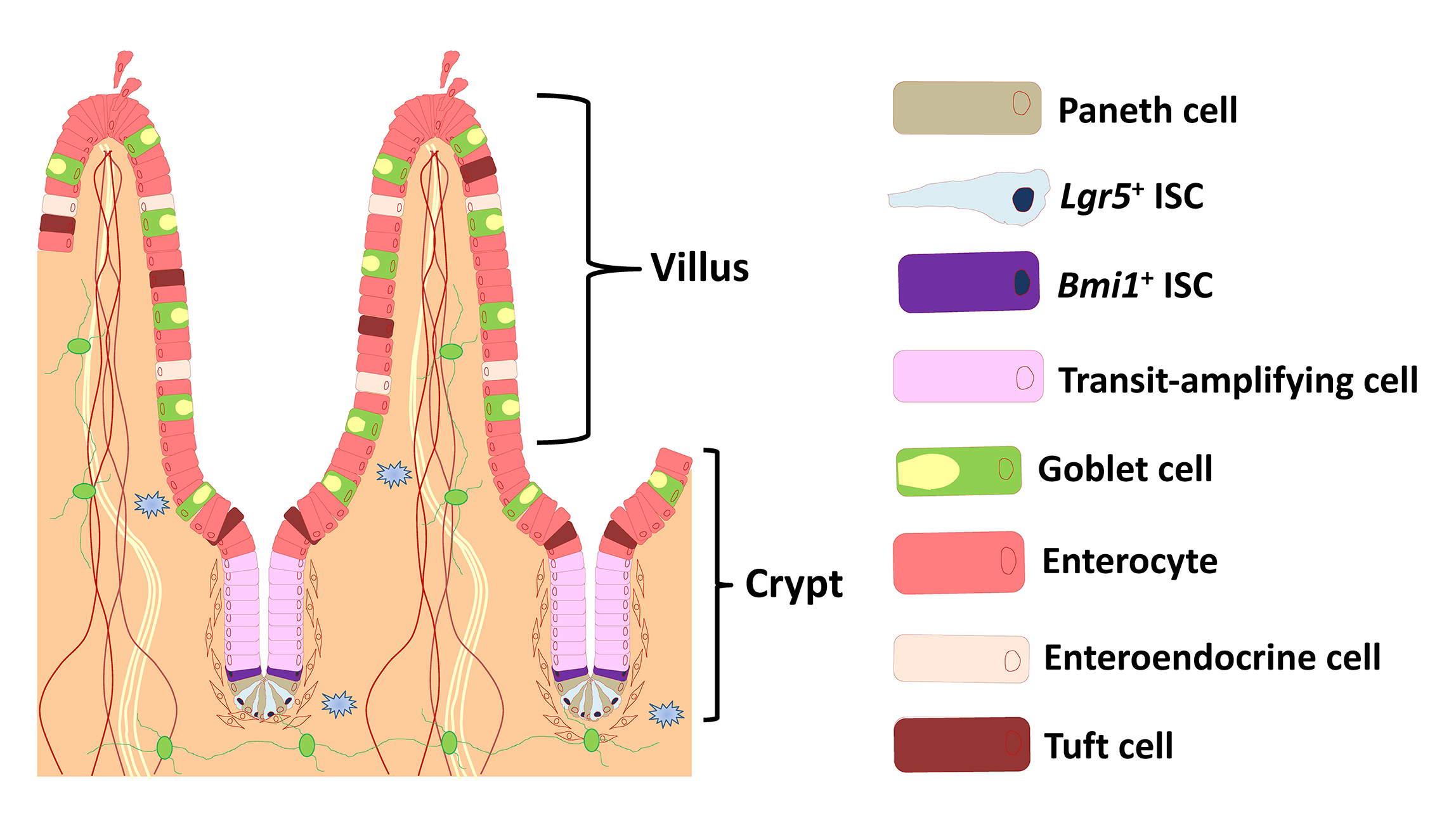
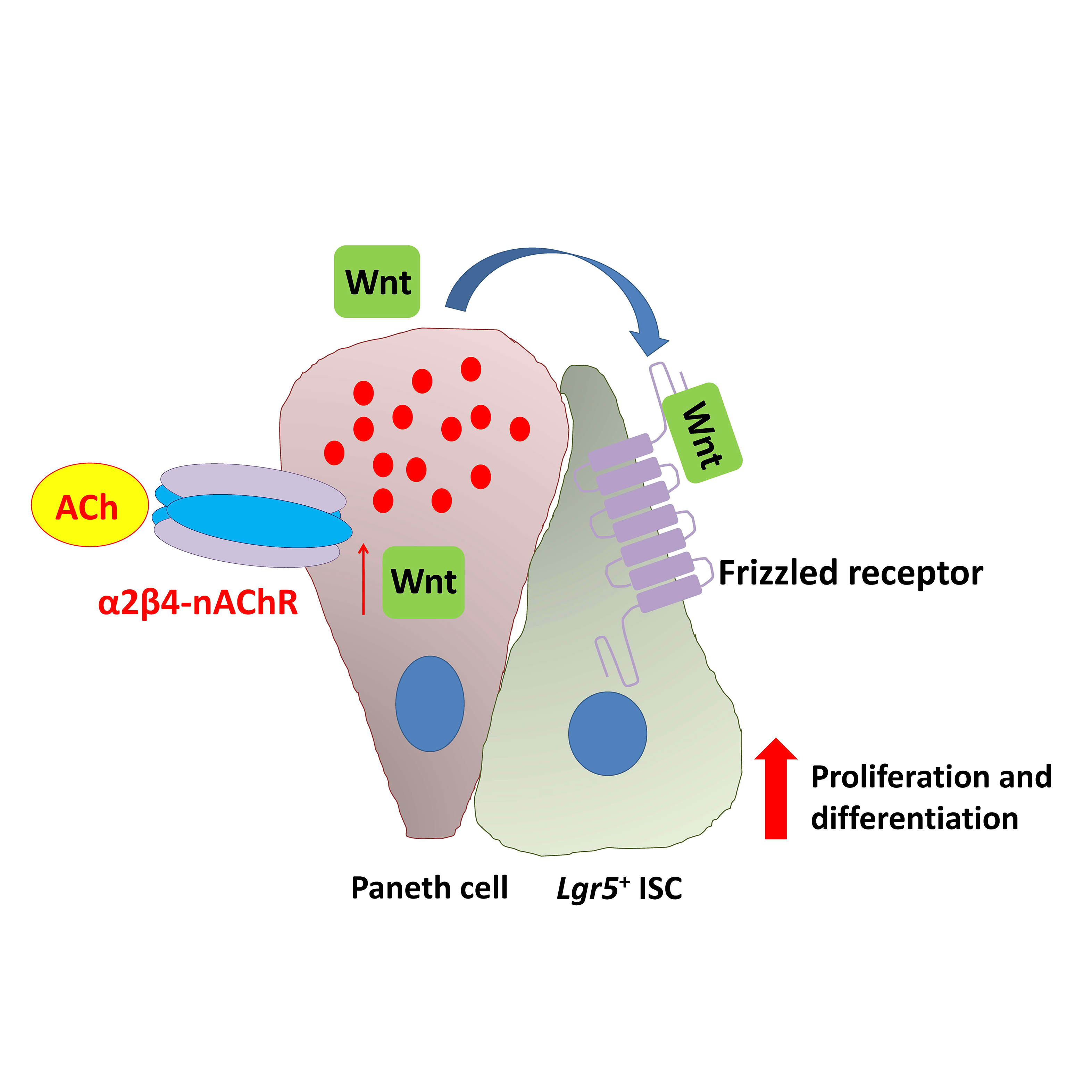
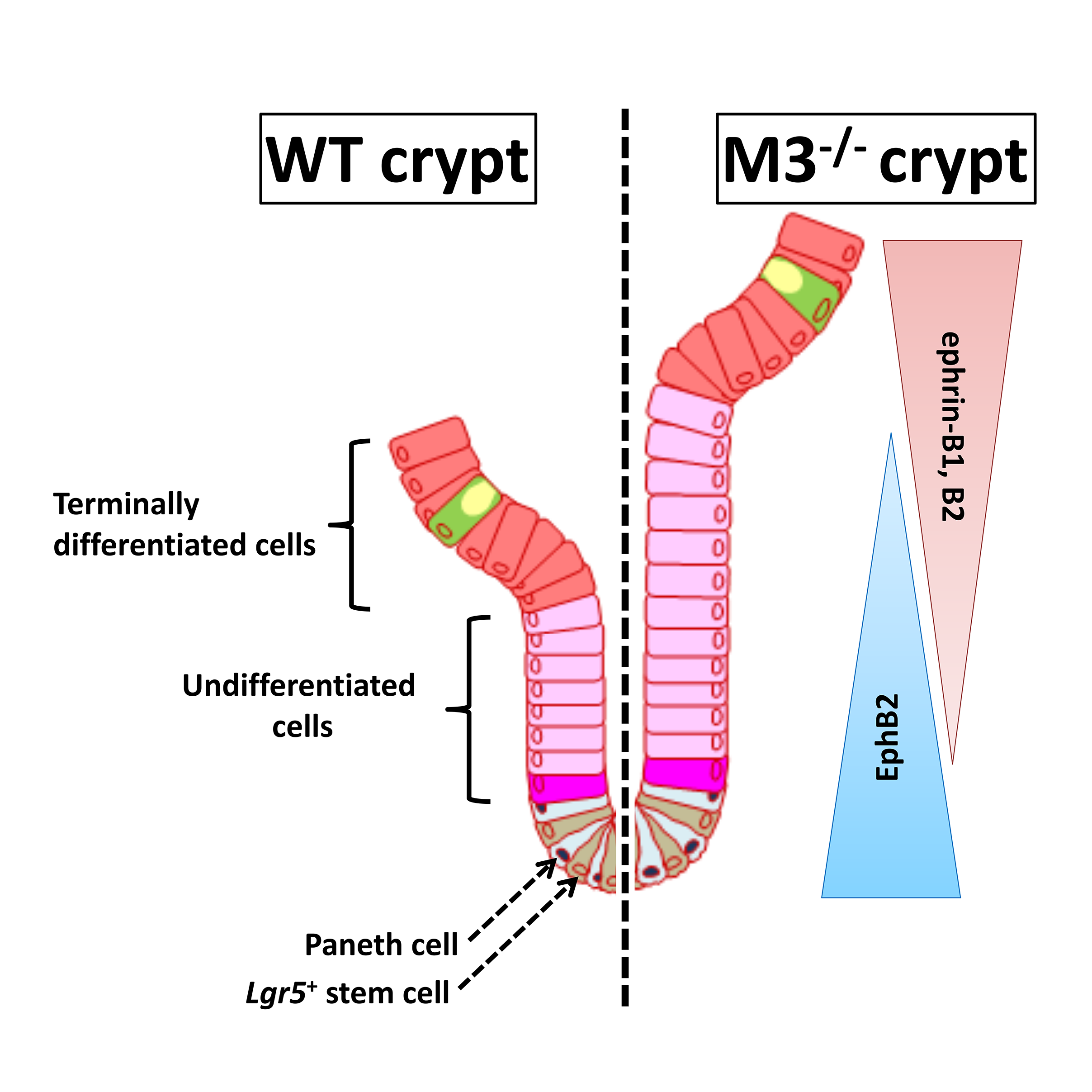
Acetylcholine (ACh) signaling through activation of nicotinic and muscarinic ACh receptors (nAChRs and mAChRs) regulates expression of specific genes that mediate and sustain proliferation, differentiation, and homeostasis in the intestinal crypts. This signaling plays a pivotal role in the regulation of ISC function, but the details have not been clarified. We have revealed at the first time that α2β4-nAChR localized at Paneth cells regulates ISC proliferation and differentiation coordinated with Wnt signaling (Fig. 2). Moreover, we have shown that M3-mAChR signaling coordinates the EphB/ephrin-B signaling cascade to sustain intestinal epithelial homeostasis, which in part plays a role in regulating the rate of proliferation of epithelial cells (Fig. 3).
Our findings suggest that signaling through the M3-mAChR and α2β4-nAChR appears to work together to maintain the homeostasis of intestinal epithelial growth and differentiation following modifications of the cholinergic intestinal niche. To verify the hypothesis, we have currently been carrying out systematic analyses of the cholinergic signaling pathways underlying ISC maintenance.
3. Elucidation of the new biological roles and possible applications of phytosiderophores in plants and mammals
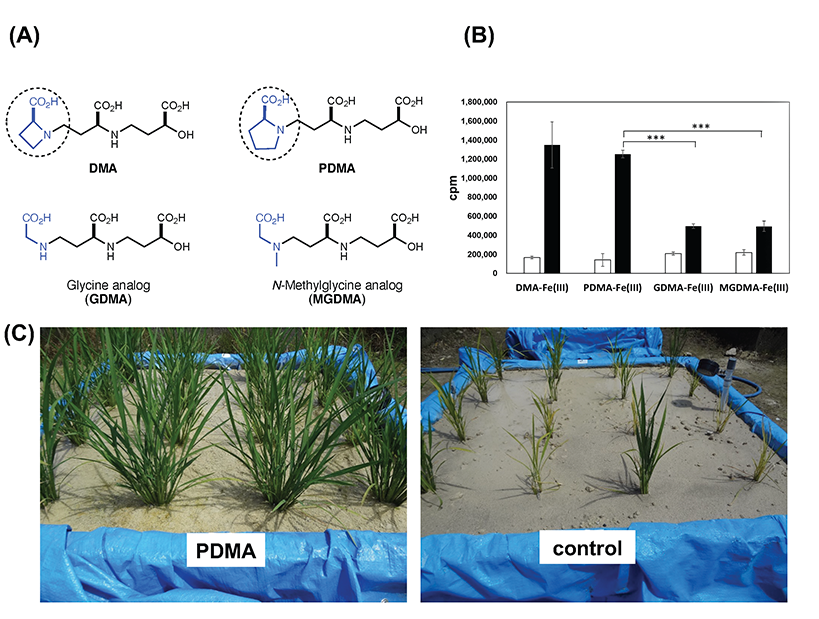
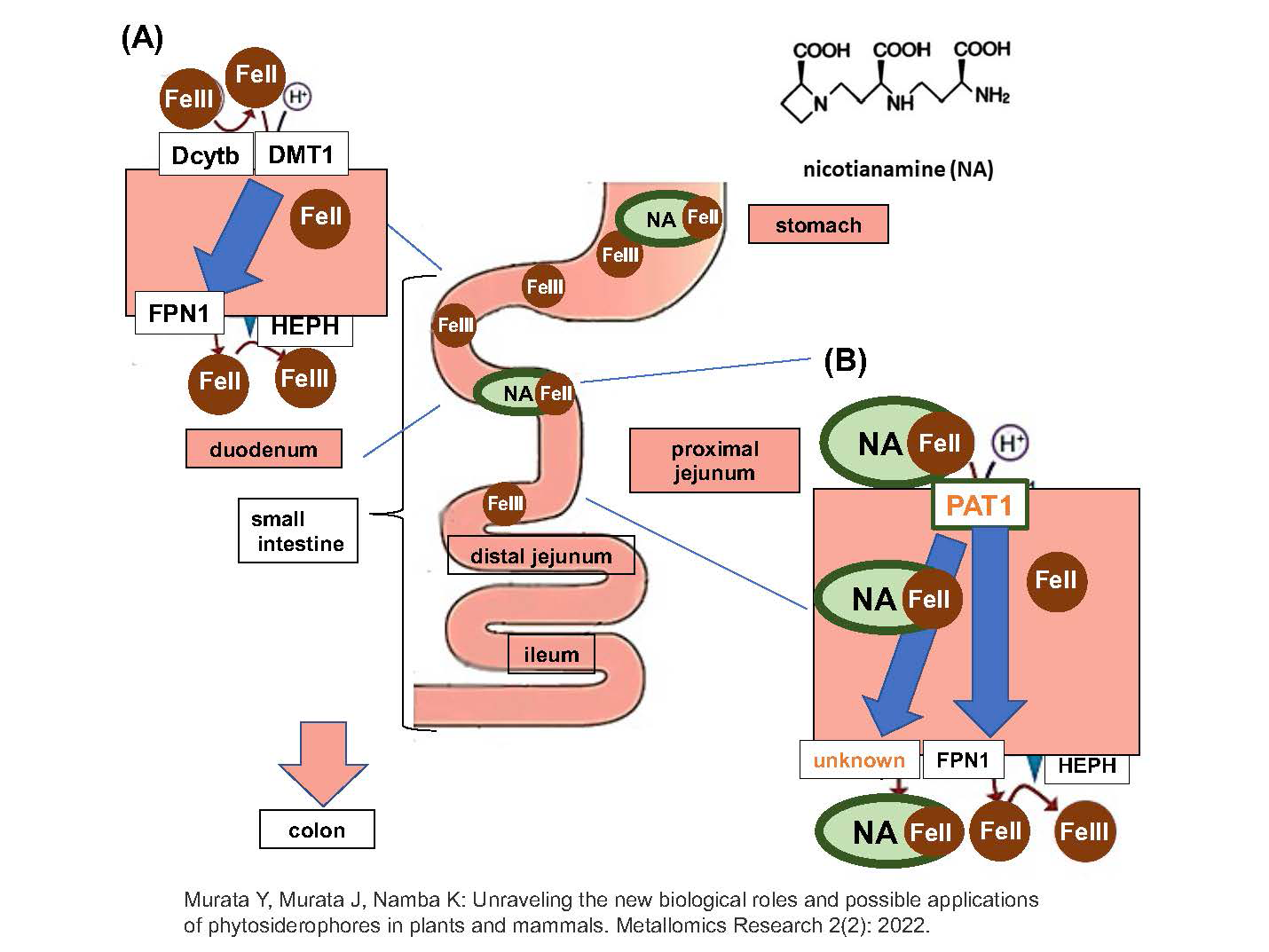
Iron is an essential element for all living organisms. The ability of plants to absorb inorganic iron from the soil is crucial for plants and mammals, which ultimately rely on plants as their nutrient source. In contrast to most plant species, Poaceae plants, including rice, have developed a distinct chelation strategy to efficiently acquire insoluble soil iron using iron-chelating substances such as mugineic acid (MAs), called phytosiderophores. Genes involved in the biosynthesis and transport of MAs and their resulting iron(III)-MA complexes across membranes have been identified. On the other hand, an efficient short-step synthesis of the substrates MA and 2′-deoxymugineic acid (DMA) has enabled a sufficient supply of these compounds. Furthermore, a synthetic analog, proline-2′-deoxymugineic acid (PDMA), which is a cost-effective MA with sufficient phytosiderophore activity was shown to promote rice growth in alkaline soil at an experimental field scale (Fig. 1).
Nicotianamine (NA), a precursor of MAs, which is essential for metal translocation within plant tissues, was recently shown to be absorbed as an iron(II) complex in the mouse small intestine by an amino acid transporter (Fig. 2). The discovery of the biological role of NA in iron absorption at the small intestine highlights the biological significance of NA crossing the plant and animal kingdoms, thus opening up new possibilities for biofortification approaches.
4. Decoding a novel mode of chemical communication between plants and soil microorganisms
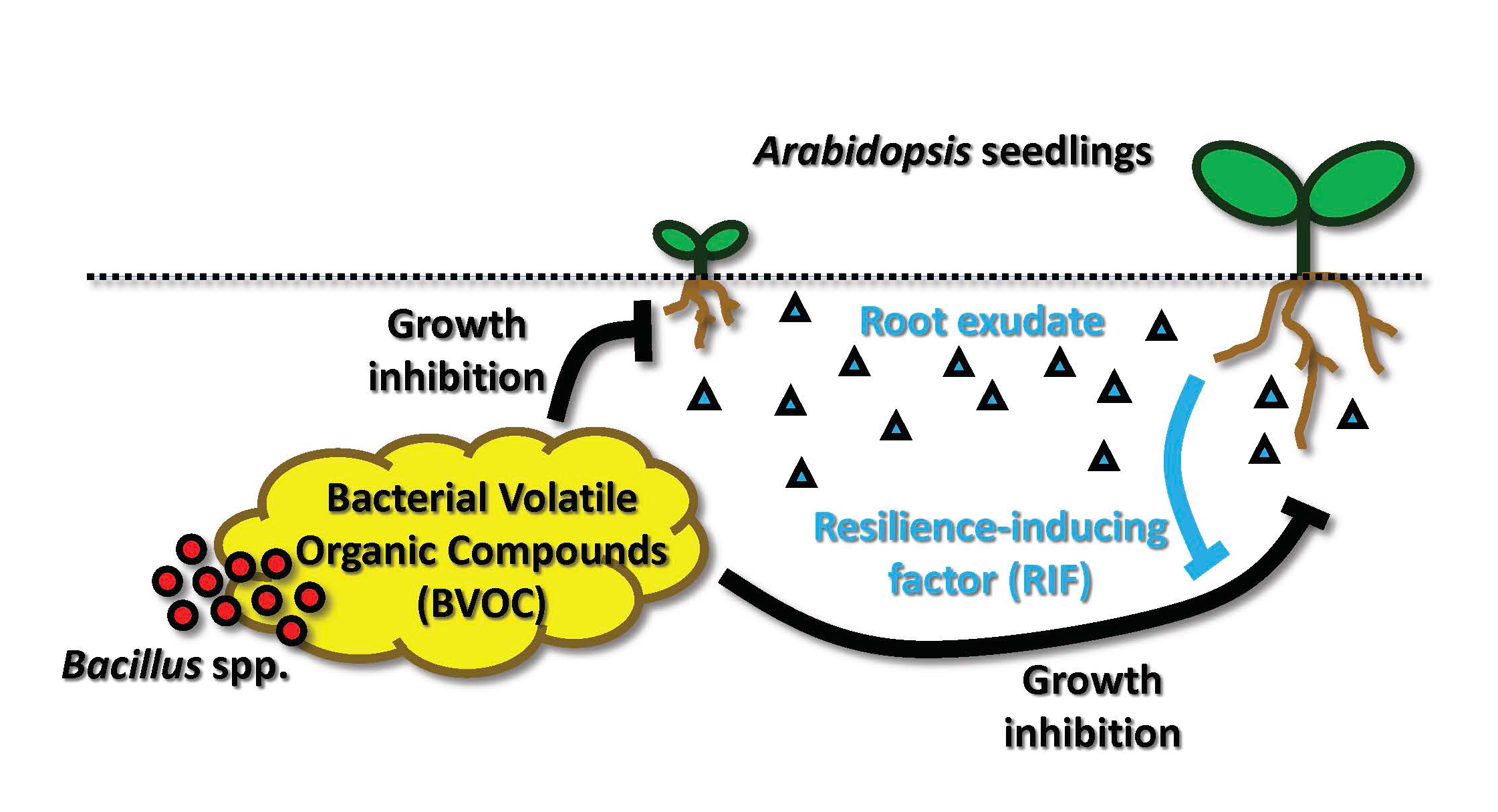
Plants inevitably encounter a wide variety of microorganisms in the rhizosphere. Recent studies demonstrate that both plant roots and soil microorganisms emit and/or secrete a spectrum of small molecules and use them to mutually affect their fitness for survival. However, the molecular mechanisms behind the interactions of plants and soil microorganisms have been characterized primarily with selected pathogens, Rhizobium, and AM fungi, and whether and how plants respond to most of the rest of soil microorganisms remains largely elusive.
Using Arabidopsis thaliana (thale cress) and Bacillus spp. as model organisms, we study (i) how bacterial volatile organic compounds (BVOC) produced by soil microorganisms affect plant growth, and (ii) how plants respond and adapt to such a stressful environment when exposed to BVOC. We found that BVOC emitted from Bacillus spp. can interfere the growth of Arabidopsis seedlings and performed bioassay-guided fractionation of BVOC for identifying the key chemical compounds that are responsible for such inhibition of plant growth. Furthermore, we observed a unique behavior of Arabidopsis seedlings upon exposure to BVOC; the seedlings communicate each other via root exudates to increase their fitness under such stressful environment. We call this novel activity that counteracts the growth inhibitory activity of BVOC as ‘resilience-inducing factor (RIF)’ and have been trying to identify the active components from root exudates.
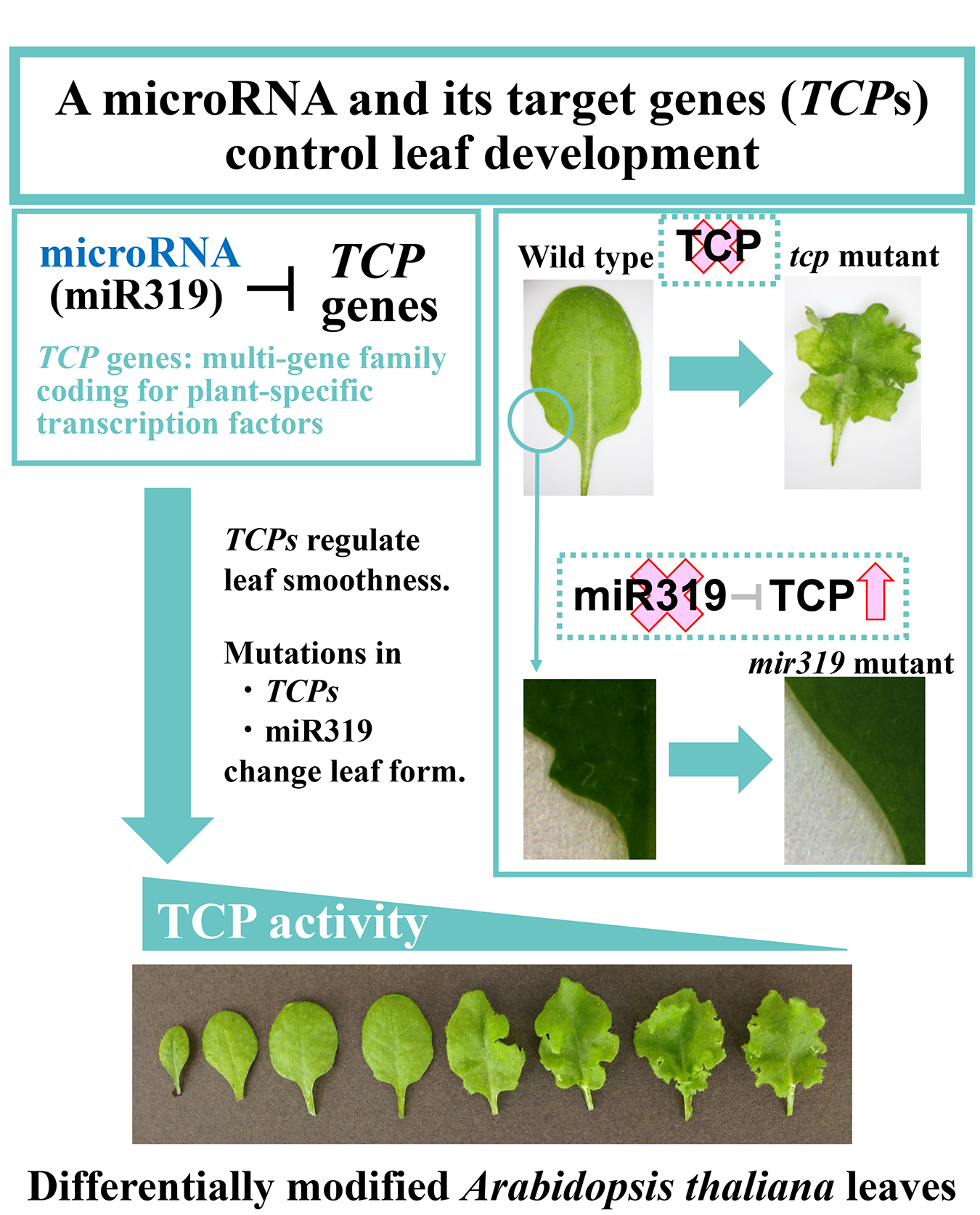
5. Analysis of the molecular mechanism of leaf development
Plants generate oxygen, fix carbon-the source of diets and fuel-and produce medicinal substances, all of which we need for our survival. Plants also provide comfort and beauty in our daily life and contribute to our high-quality lifestyle. We cannot live without plants and, therefore, must explore better ways to preserve and improve existing plants for future generations. This scientific research mainly aims to obtain a mechanistic understanding of plant development and provide technical advances in plant breeding and biotechnology to establish a future sustainable lifestyle.
Plants develop through the coordination among many processes of differentiation, morphogenesis, physiology, metabolism, senescence, and responses to environmental stress. This coordination is achieved by master regulators of plant development. Because these regulators often play pleiotropic roles in a plant’s life cycle, this research project focuses on the developmental regulation mechanism of a leaf as a model organ. A major goal of this project is to provide a framework for the coordination among individual processes in leaf development.
This project currently conducts experiments to understand the molecular mechanisms of leaf development. It focuses on the signaling pathway downstream of the microRNA-regulated transcription factors and the accumulation of metabolites. Emphasis is also placed on the genetic modification of leaves with a novel morphology.